back to summary
-
You can access the necessary files for this tutorial by downloading this
archive. Once downloaded, place it in a convenient location and extract its contents:
[user]$ tar -xvjf 8_inductive.tbz2
Unpacking the archive will create a directory called 8_inductive/, wherein this tutorial will be conducted. - The results of the calculations can be found in this archive_results.
VIII. Inductive attractive electronic effect
Go to directory 8_inductive/In order to provide users with a simple tool to assess inductive effects on specific bonds in molecules, the new Pair Density Asymmetry (PDA) index has been devised.
In this section, we are going to assess the electronic effect of one, two and three fluorine atoms on the central C-C bond of ethane.
1) The reference molecule: C2H6
Change to directory 0_REFThe wave function is stored in the file mol.wfx.
Edit and save the following param.igm input file (the file param.igm must be located in the same directory as the mol.wfx file supplied for you):
1
mol.wfx
IBSI
1 5
ENDIBSI
- 1 --> one single file describes the system
- mol.wfx --> the name of this file --> here, the electron density (ED) will be calculated from a wave function
(note the the old wfn file format is also supported by IGMPlot) - IBSI/ENDIBSI: keyword to start/end the IBSI section
- 1 5: the C1 - C5 atom pair is examined
The calculation of the PDA index is performed within a IBSI calculation (bond strength). The calculation takes a few seconds (depending on the core numbers).
Provided that you have installed IGMPlot (see documentation for installation instructions): run the program IGMPlot with the command in a linux terminal:
-> IGMPLOT param.igm > igm.log &
Edit the resulting igm.log file and look at the "FINAL BONDING FEATURES". The Pair Density Assymmetry for the central C-C bond is 0.0:
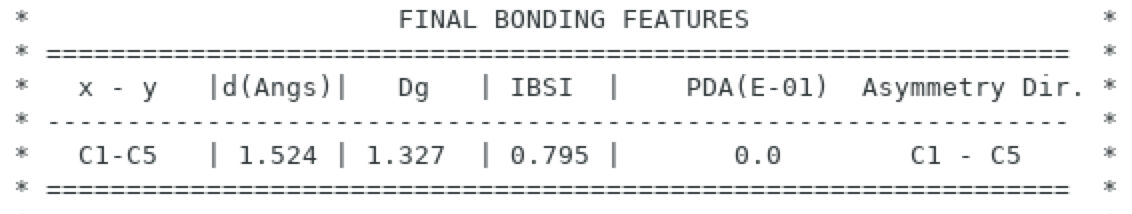
This results (no asymmetry) makes sense since the electron distribution in this molecule is totally symmetrical.
2) One substitution: H3C-CH2F
Change to directory 1_1FWe are now going to measure the electronic effect of one fluorine atom on the electronic distribution of the central C-C bond.
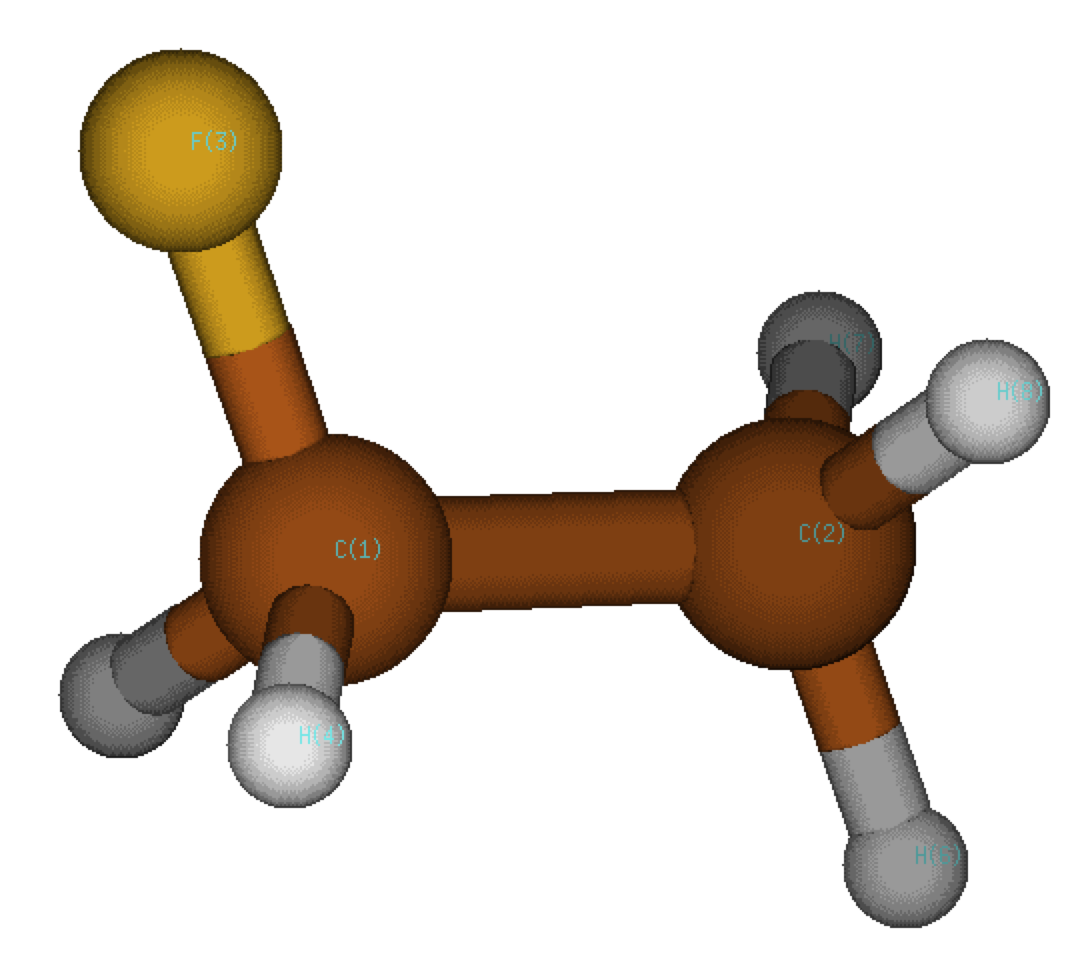
The wave function is stored in the file mol.wfx.
Edit and save the following param.igm input file (the file param.igm must be located in the same directory as the mol.wfx file supplied for you):
1
mol.wfx
IBSI
1 2
ENDIBSI
- 1 --> one single file describes the system
- mol.wfx --> the name of this file --> here, the electron density (ED) will be calculated from a wave function
(note the the old wfn file format is also supported by IGMPlot) - IBSI/ENDIBSI: keyword to start/end the IBSI section
- 1 2: the C1 - C2 atom pair is examined
run the program IGMPlot with the command in a linux terminal:
-> IGMPLOT param.igm > igm.log &
The calculation may take a few seconds. The resulting PDA value is now: 0.45:
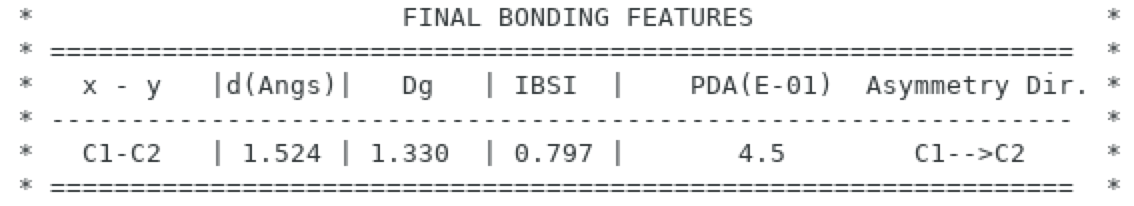
So, the presence of a fluorine atom asymmetrically distorts the electron density of the central C-C bond in the direction of the carbon atom 2 without the fluorine. Clearly, having one fluorine atom on only one of the two carbon atoms generates a slight asymetry, which is measured here through the PDA index. To give an idea, the asymmetry of a C-F bond is about 10. So, the effect of one single fluorine atom represents only about 4% of the C-F asymetry. Furthermore, the direction of asymetry points towards the carbon 2.
3) Two substitutions: H3C-CHF2
Change to directory 2_2FThe wave function is stored in the file mol.wfx.
Edit and save the following param.igm input file (the file param.igm must be located in the same directory as the mol.wfx file supplied for you):
1
mol.wfx
IBSI
1 2
ENDIBSI
run the program IGMPlot. The resulting PDA value is now: 8.2.
4) Three substitutions: H3C-CF3
Change to directory 2_2FThe wave function is stored in the file mol.wfx.
Edit and save the following param.igm input file (the file param.igm must be located in the same directory as the mol.wfx file supplied for you):
1
mol.wfx
IBSI
1 2
ENDIBSI
run the program IGMPlot. The resulting PDA value is now: 11.5.
5) Summary
| 0F | 1F | 2F | 3F | |
| PDA (10-1) | 0.0 | 4.5 | 8.2 | 11.5 |
The PDA is sensitive to the chemical surroundings of the atom pair. That is the whole point of the PDA tool: to examine and quantify the effect of external factors on a given bond. The electronic effect caused by one, two or three fluorine atoms on the adjacent C–C bond of ethane initially purely symmetrical is well described here.
In a homonuclear diatomic molecule, in the absence of external perturbation, electrons are equally distributed between the two atoms leading to a symmetrical picture of the electron density (ED) along the bond. In contrast, when different atoms bond together the ED is accumulated unequally between atoms. The PDA index gives a measure of the ED asymmetry in between the two atoms and the direction of the asymmetry.
The PDA index takes the asymmetry information from the electron density gradient (see the documentation for more details). The direction of asymmetry as measured by the PDA points towards the atom bringing the most electrons in the bond axis (z) direction. Three factors govern the magnitude and direction of the PDA, in descending order:
- the period of each atom decides of the resulting direction and magnitude for the PDA. For instance, the PDA value of C−H bond in CH4 (22.9) is much larger than the PDA of C–F (10.6). This makes sense since in CH4, the 1s atomic core orbital of C brings large electron density gradient in the bond axis direction (among others) while H has no core counterpart
- the electronegativity difference between the two atoms of the considered pair plays a major role
- the electronic environment of the studied bond can influence the PDA of this bond
Noteworthy, the PDA analysis is exclusively restricted to the region in between the examined atom pair and is tackled directly at the gradient level. It has nothing to do with the bond polarity derived from a conventional point charge analysis. Actually, the latter is not based on the ED gradient (but rather on the ED and nuclear charges) and it incorporates, to some extents, the ED distribution asymetry (whenever there is) of all neighbors beyond the examined atom pair. However, both analyses (PDA and partial atomic charges) can be complementary tools to probe the ED distribution and the effect on it caused by perturbations.